REVIEW ARTICLE
ADAM and ADAMTS Proteases in Hepatic Disorders
Julia Bolik1, Janina E. E. Tirnitz-Parker2,3, Dirk Schmidt-Arras1
1Institute of Biochemistry, Christian-Albrechts-University Kiel, Kiel, Germany; 2School of Pharmacy and Biomedical Sciences, Curtin Health Innovation Research Institute, Curtin University, Perth, Australia; 3School of Biomedical Sciences, University of Western Australia, Perth, Australia
Abstract
Proteolysis is an irreversible post-translational modification that regulates protein function and signal transduction. This includes remodelling of the extracellular matrix, release of membrane-bound cytokines and receptor ectodomains, as well as the initiation of intracellular signalling cues. Members of the adamalysin protease subfamily, in particular the ADAM (a disintegrin and metalloprotease) and ADAMTS (the ADAM containing thrombospondin motif) families, are involved in these processes. This review presents an overview of how ADAM and ADAMTS proteins are involved in liver physiology and pathophysiology.
Keywords: ADAM; ADAMTS; metzincin superfamily; thrombotic thrombocytopenia purpura; von Willebrand Factor
Received: 10 December 2018;
Accepted after revision: 17 January 2019;
Published: 07 February 2019
Author for correspondence: Dirk Schmidt-Arras, Christian-Albrechts-University Kiel, Institute of Biochemistry, Kiel, Germany. Email:
darras@biochem.uni-kiel.deHow to cite: Bolik J. et al. ADAM and ADAMTS proteases in hepatic disorders. J Ren Hepat Disord. 2019;3(1):23–32
Doi: http://dx.doi.org/10.15586/jrenhep.2019.47Copyright: Bolik J. et al.
License: This open access article is licensed under Creative Commons Attribution 4.0 International (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0
Introduction
The liver harbours different cell types, including hepatocytes, cholangiocytes, resident macrophages called Kupffer cells (KCs), sinusoidal endothelial cells (SECs) and hepatic stellate cells (HSCs). Hepatocytes, the liver parenchymal cells, make up the vast majority of cells and fulfil multiple vital body functions such as protein synthesis and storage; detoxification; synthesis of cholesterol, phospholipids and bile salts; and secretion of bile. Bile is then stored in the gallbladder and drained through bile ducts formed by cholangiocytes to aid in the digestion and absorption of dietary fats and fat-soluble vitamins in the duodenum. The liver sinusoids are lined with KCs and SECs, while HSCs represent perisinusoidal cells found in the space of Disse, an area between liver SECs and hepatocytes. In homeostatic conditions, HSCs store fat and fat-soluble vitamins in the liver, in particular vitamin A (retinol, retinoic acid).
Upon acute liver damage, hepatocytes restore the lost liver mass by proliferation and hypertrophy (1). However, under chronic toxic, viral or carcinogenic insult, the proliferation of hepatocytes is inhibited, and they often become senescent. Under these circumstances, hepatic progenitor cells (HPCs) are activated and observed to proliferate in a variety of chronic liver diseases, including alcoholic liver disease, non-alcoholic fatty liver disease, steatohepatitis and in the iron overload disorder, haemochromatosis. HPCs are small, stem-cell-like cells with unclear origin and have the capacity to differentiate into hepatocytes or cholangiocytes, depending on the underlying injury stimulus (2). Damage signals from cellular compartments lead to the transdifferentiation of HSCs to proliferative and fibrogenic myofibroblasts. These “activated” HSCs respond by depositing collagen, resulting in the formation of scar tissue, which may progress to liver cirrhosis in severe cases. Numerous studies have reported a close temporal and spatial organisation of KCs, HPCs and HSCs, which display an intimate interplay and orchestrate liver regeneration versus disease progression through cytokine and chemokine cross-talk (2–6). These complex signalling networks require precise regulations not only at the cellular level but also at the molecular level. Members of the metzincin protease family have been shown to be involved in different aspects of chronic liver disease and tumour formation.
The Metzincin Superfamily
The superfamily of zinc proteases or metzincins is characterised by the presence of a protease domain containing an invariant HExxHxxGxxH zinc-binding motif (7, 8). It is subdivided into four subfamilies: matrixins, adamalysins, astacins and bacterial serralysins. The SVMPs (snake venom metalloproteinases), the ADAMS (a disintegrin and metalloproteinases) and ADAMTs (ADAMs containing thrombospondin motifs) form the adamalysin subfamily (9). The catalytic domains of metzincins share a similar overall structure with the catalytic cleft positioned between an N-terminal subdomain (NSD) and a C-terminal subdomain (CSD) (8). The NSD is anchored by a five-stranded β sheet, followed by a central α helix which contains the HExxH motif supplying two of the histidines involved in Zn2+-coordination and the glutamate residue that participates in catalysis (Figure 1). C-terminal to the α-helix, a conserved methionine turn is packed against the zinc-binding site (8, 10). Both ADAM and ADAMTS protein structures are thought to exist as “open” or “closed” structures representing another layer of regulation. Currently, this has been shown for the members ADAM17 and ADAMTS4 and 5, respectively (11, 12). In the case of the closed ADAMTS4 conformation, movement of the S2’ loop towards the catalytic centre precludes substrate binding and leads to the release of an additional Ca2+ ion (11).
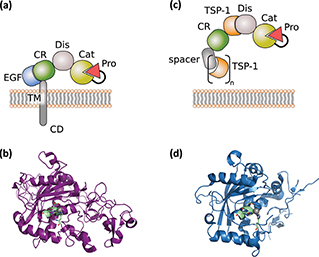
Figure 1. General structure of ADAM and ADAMTS proteases. (a) General domain structure of ADAM proteases. (b) X-ray structure (6BE6.pdb) of ADAM10, including catalytic, disintegrin and cysteine-rich domains. The catalytic zinc is highlighted in grey and the zinc-binding motif in pale green. (c) General domain structure of ADAMTS proteases. (d) X-ray structure of ADAMTS1 (2V4B.pdb), including catalytic and disintegrin domain. The catalytic zinc is highlighted in grey and the zinc-binding motif in pale green.
The ADAM Family
In mammals, ADAMs are expressed in a wide range of tissues. Given their numerous substrates, they have diverse functions in development, physiology and pathology (9). The human genome encodes for 22 functional ADAM proteins, of which 10 are considered proteolytically inactive (13). ADAMs without protease activity are thought to facilitate protein folding and protein–protein interactions.
The domain structure of ADAM proteases comprises an N-terminal inhibitory pro-domain, a catalytic metalloprotease domain, a disintegrin domain with a cysteine-rich region, an epidermal growth factor (EGF)-like domain, a transmembrane domain and a cytoplasmic tail (Figure 1a). ADAM proteases are synthesised as catalytically inactive transmembrane proteins of about 750 amino-acid length into the endoplasmic reticulum. The N-terminal pro-domain is thought to have chaperone and inhibitory functions as it interferes with the Zn2+-ion in the catalytic centre. Within the Golgi apparatus, ADAMs undergo further complex glycosylation and are subjected to proteolytic cleavage by the Furin protease, thereby liberating the N-terminal pro-domain. However, for ADAM8 and ADAM28, autocatalytic pro-domain cleavage was demonstrated (14, 15). Recently it has been shown that the recombinant pro-domain of ADAM17 can be harnessed as a specific inhibitor in vitro and in vivo (16).
The catalytic domain is conserved among the ADAM family members and contains the zinc-binding motif (HEx-GHxxGxxHD) (Figure 1 a and b) (17). The adjacent disintegrin and cysteine-rich domains are suggested to be involved in autoregulation as they fold back and limit access to the catalytic site in the unliganded state (10, 18). The disintegrin domain may also participate in cell–cell adhesion processes and has been shown for ADAM10 to play a role in substrate recognition in concert with the cysteine-rich domain (9, 19). The cytoplasmic tails of transmembrane ADAMs contain phosphorylation sites for several kinases, denoting a role in regulation of protease activity and downstream signalling (9). ADAM-mediated shedding can either be constitutive or activated by G-protein-coupled receptors; Ser/Thr-kinase activity, including protein kinase C (PKC), ERK and p38; and increased intracellular Ca2+(9, 20, 21).
ADAM proteases play a major role in proteolytic ectodomain cleavage, a process termed “shedding” (Figure 2). Functionally diverse proteins have been shown to be subjected to ectodomain shedding with differential physiological consequences. Shedding of receptor molecules such as transforming growth factor (TGF) β receptor abrogates its signalling. Importantly, the soluble ectodomain has the capacity to work as a scavenger and binds the corresponding cytokine (22). The receptor for interleukin 6 (IL-6) is an exception because the IL-6/soluble IL-6R (sIL-6R) complex induces signalling via gp130 on target cells. This process has been termed “IL-6 trans-signalling.” (23, 24)
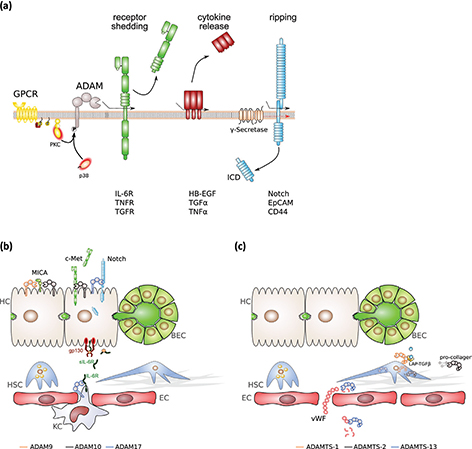
Figure 2. Signal activities of ADAM and ADAMTS proteinases in the liver. (a) ADAM protease activity, ADAM17 in particular, can be regulated by protein phosphorylation. Active ADAM proteases are involved in the release of receptor ectodomains, thereby blunting receptor signalling in most of the cases, the release of membrane-bound cytokines and the initiation of intracellular signalling by regulated intramembrane proteolysis (RIP). (b) Selected proteolytic events of the indicated ADAM proteinases in different cell types of the liver. (c) Selected proteolytic and non-proteolytic events of the indicated ADAMTS proteinases in different hepatic cell types. BEC, biliary epithelial cells; EC, endothelial cell; gp130, glycoprotein 130; HSC, hepatic stellate cells; HC, hepatocyte; ICD, intracellular domain; IL-6R, interleukin 6 receptor; KC, Kupffer cell; LAP-TGFβ, latency-associated peptide-transforming growth factor β; MICA, MHC class I polypeptide-related sequence A; sIL-6R, soluble IL-6R; vWF – von Willebrand factor.
Membrane-bound cytokines and growth factors are also liberated from the membrane by ectodomain shedding (Figure 2). The most prominent example is the family of EGF ligands, comprising heparin-binding EGF (HB-EGF), TGFα, epiregulin and neuregulin, which are shed by ADAM17, and known to play key roles in tumour growth (25). The most prominent substrate of ADAM10 is the Notch receptor, which has been implicated in developmental processes, stem cell growth and differentiation (26). In addition, there is evidence that ADAMs are able to degrade extracellular matrix (ECM) components: ADAM10 cleaves type IV collagen; ADAM13 and ADAM9 degrade fibronectin (27).
ADAM9
ADAM9 processes a wide range of substrate molecules, including amyloid precursor protein (APP), collagen XVII and HB-EGF, and has been linked to cell proliferation, adhesion and migration (28).
Transcriptomic analysis of fibrotic liver tissue revealed that expression of ADAM9 correlated with the activation of HSCs, as assessed by quantitation of alpha-smooth muscle actin (αSMA), independent of the underlying disease aetiology. Northern blots analysis demonstrated that ADAM9 expression was localised to HSCs (Figure 2b) and that expression of ADAM9 significantly increased in activated compared to quiescent HSCs (Table 1) (29). These data suggest that ADAM9 is important for ECM remodelling during hepatic fibrosis progression and may thereby contribute to the establishment of an environment conducive to hepatocellular carcinoma (HCC). Indeed, various studies have reported ADAM9 overexpression in HCC (30–32), and high expression levels of ADAM9 have been linked to tumour aggressiveness (30). Beside its effect on HSCs, ADAM9 was found to promote ectodomain shedding of major histocompatibility class I-related chain A (Figure 2b). Consequently, HCC cells with siRNA-mediated suppression of ADAM9 were more susceptible to natural killer cell-mediated cytolysis. Interestingly, sorafenib-treatment reduced ADAM9 expression, and enhanced major histocompatibility class I-related chain A protein levels and anti-tumour response of natural killer cells (32). An alternatively spliced ADAM9 variant (ADAM9-S), secreted by activated HSCs and stromal liver myofibroblasts, has been shown to promote tumour metastasis to the liver (Table 1). Through its disintegrin domain, ADAM9-S directly binds to α6β4 and α2β1 integrins on colon carcinoma cells and is able to cleave laminin, thereby promoting tumour cell invasion (33).
Table 1. Overview of known ADAM and ADAMTS proteinase activities in the liver and their association with hepatic diseases.
| The ADAM family |
| Family member |
Associated disease |
Substrate(s) |
Cell type |
Reference |
| ADAM9 |
Fibrotic liver disease |
ECM components |
Activated HSCs |
(29, 30) |
| |
HCC |
MIC-A |
Hepatocytes |
(32) |
| |
Liver metastasis |
Laminin, binding to integrin α6β4, α2β1 |
Activated HSCs |
(33) |
| ADAM10 |
Liver homeostasis |
pot. indep. of catalytic activity |
Hepatocytes, HPCs |
(37) |
| |
Liver fibrosis |
CX3CL1 |
Activated HSCs |
(38, 39) |
| |
Murine cholestasis |
c-Met |
Hepatocytes |
(41) |
| |
HCC |
MIC-A |
Hepatocytes |
(39) |
| |
Liver metastasis |
L1CAM |
Tumour cells |
(43) |
| |
|
c-Met |
HSCs |
(44, 45) |
| ADAM12 |
Cirrhotic liver |
pot. ECM remodelling |
Activated HSCs |
(47) |
| |
HCC |
pot. ECM remodelling |
Activated HSCs |
(48) |
| ADAM17 |
Liver damage |
CX3CL1 |
Activated HSCs |
(38) |
| |
HCC |
Notch |
Activated HSCs |
(53) |
| |
|
pot. EGFR ligands |
Hepatocytes |
(50, 51) |
| |
|
pot. IL-6R |
Kupffer cells |
(56) |
| The ADAMTS family |
| Family member |
Associated disease |
Substrate(s) |
Cell type |
Reference |
| ADAMTS1 |
Fibrotic liver disease |
Binding to LAP-TGFβ |
HSCs |
(59) |
| ADAMTS2 |
Murine liver fibrosis |
Pro-collagen |
HSCs |
(62) |
| ADAMTS13 |
SAH |
vWF |
endothelium |
(71) |
| |
ALI, ALF |
|
|
(72) |
| |
Fibrotic liver disease |
|
|
(74–78) |
| ADAM, a disintegrin and metalloprotease; ADAMTS; the ADAMs containing thrombospondin motif; ECM, extracellular matrix; HCC, hepatocellular carcinoma; MIC-A, major histocompatibility class I-related chain A; HSC, hepatic stellate cell; HPC, hepatic progenitor cell; L1CAM, L1 cell adhesion molecule; IL-6R, interleukin 6 receptor; EGFR, epidermal growth factor receptor; LAP-TGFβ, latency-associated peptide-transforming growth factor β; SAH, severe alcoholic hepatitis; ALI, acute liver injury; ALF, acute liver failure; vWF, von Willebrand Factor. |
ADAM10
In most tissues investigated, ADAM10 is the major protease that initiates Notch signalling. In the liver, biliary tree formation (34, 35) and differentiation of HPCs into cholangiocytes (4) are dependent on Notch signalling (Figure 2a). This is also reflected by the finding that patients with Alagille syndrome who suffer from biliary paucity and subsequent cholestasis display mutations in the Notch ligand Jagged-1 (36). We recently investigated the role of ADAM10 under physiological conditions by generating liver-specific ADAM10-deficient mice. Surprisingly, we observed that ADAM10 was dispensable for Notch2 activation in vitro in an HPC line, and for biliary tree formation in vivo (37), further suggesting that during development, Notch signalling in hepatoblasts does not rely on ADAM10. However, we observed that hepatic loss of ADAM10 leads to the down-regulation of biliary transporters, resulting in spontaneous hepatocyte necrosis (Table 1). Furthermore, loss of ADAM10 resulted in an accumulation of HPCs that might at least, in part, be the consequence of enhanced signalling through the hepatocyte growth factor receptor c-Met (37).
Overexpression of ADAM10 has been reported in chronic liver disease associated with liver fibrosis (38) and in HCC (39, 40) (Table 1). In HSCs, ADAM10 is involved in the release of the soluble chemokine, CX3CL1, thereby facilitating recruitment of inflammatory cells (38). Furthermore, c-Met ectodomain release by ADAM10 from HSCs correlated with hepatic injury in the murine 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) cholestasis model (41). Soluble c-Met has previously been identified as a decoy receptor and might therefore restrict the proliferative response after liver damage (42). SiRNA-mediated suppression of ADAM10 in the human liver cancer cell line, HepG2, decreased cellular proliferation, the ability to grow in semi-solid medium and its tumorigenic potential in xenografts (39, 40). ADAM10 might also be involved in anti-tumour immunity as it has been demonstrated that major histocompatibility class I-related chain A is a substrate for ADAM10 in HepG2 cells (Figure 2b, Table 1), where its surface localisation was enhanced in the absence of ADAM10 (39). Inhibition of ADAM10 might therefore represent an attractive avenue to increase the anti-tumour immune response.
In addition to its role in fibrotic liver disease and HCC, ADAM10 seems to have a role in the establishment of liver metastasis. The neuronal cell adhesion receptor L1-CAM, which is expressed on colon cancer cells, is proteolytically processed by ADAM10, generating a L1-CAM intracellular domain (ICD). An increased L1-CAM ICD formation through enhanced ADAM10 activity resulted in enhanced liver metastasis (Table 1) (43). Furthermore, tumour cell-secreted tissue inhibitor of metalloproteinase (TIMP) 1 was shown to inhibit ADAM10-mediated c-Met processing (Figure 2b) and thereby c-Met signalling in liver metastasis. Suppression of TIMP-1 lowered the metastatic potential of tumour cells, while circulating TIMP-1 levels correlated with an increased risk for liver metastasis formation (44, 45). Very recently, the same group demonstrated that TIMP-1 secreted from pancreatic premalignant lesions activates HSCs in the liver via binding to the tetraspanin CD63, thereby preparing a hepatic premetastatic niche (46).
ADAM12
Due to alternative splicing, ADAM12 can be expressed as a long (ADAM12L) or as a short (ADAM12S) form. The latter is soluble due to its lack of transmembrane and cytoplasmic domains (47). Both ADAM12 isoforms were found to be up-regulated upon TGFβ stimulation in activated HSCs but not in hepatocytes. Furthermore, expression of ADAM12 was increased in cirrhotic livers and liver cancer and localised to cells of the tumour stroma, presumably HSCs (Table 1) (30, 48). ADAM12 was also detectable in circulation and correlated with overall survival (48). Interestingly, in another study, overall patient survival and ADAM12 expression correlated with the expression of the Tetraspanin Tspan8 (49), which is often up-regulated in HCC (50). Down-regulation of Tspan8 in HCC lines reduced ADAM12L expression and tumourigenicity in a mouse xenograft model (49). However, more in-depth analysis is needed to identify definite substrates of ADAM12 and decipher its potential role in tumour stroma dynamics and ECM remodelling.
ADAM17
Reports using in vitro experiments demonstrated that ADAM17 mediates the release of EGFR ligands in hepatocytes upon exposure to tumour necrosis factor (TNF) α (51) or TGFβ (52), suggesting that ADAM17 on hepatocytes might be involved in liver regeneration (Table 1). Furthermore, release of CX3CL1 from HSCs was also shown to be mediated by ADAM17, correlating with enhanced inflammatory cell infiltration upon liver damage (38). However, a clear in vivo evidence for a role of ADAM17 in liver regeneration is still lacking.
Overexpression of ADAM17 has been reported in human HCC and diethylnitrosamine (DEN)-induced murine HCC model (53). In tumour cells, ADAM17 is essential for Notch signalling (Figure 2b), resulting in the maintenance of a cancer stem cell phenotype (54) with enhanced migratory abilities through activation of integrin β1 (53, 55). Interestingly, it was shown that formation of hepatic metastases is enhanced by the release of soluble Notch ligands from endothelial cells, which in turn activates Notch on colorectal cancer cells. In vitro experiments using small molecule inhibitors suggested that proteolytic release of Notch ligands might be mediated by ADAM17 (56). However, the question whether ADAM17 is also important for Notch signalling under physiological conditions, in particular in the liver, remains unanswered.
ADAM17 is a major protease for the membrane-bound IL-6R, leading to the release of soluble IL-6R (Figure 2b). In contrast to many other soluble receptors, sIL-6R is able to bind IL-6 and induce signalling through the signal-transducing subunit, gp130 (24). This process is called IL-6 trans-signalling. We recently demonstrated that IL-6 trans-signalling is critically involved in HCC tumour initiation and tumour angiogenesis (56). Taken together, liver tumourigenesis appears to depend on ADAM17 proteolysis. However, more detailed in vivo analyses are needed to clarify if ADAM17 can be harnessed as a potential target in HCC therapy.
The ADAMTS Family
Unlike ADAM family proteins, members of the ADAMTS protease family contain neither transmembrane nor cytoplasmic domains and are synthesised as extracellular proteins. Instead, all family members contain at least one thrombospondin (TSP) type-1 repeat (TSR) that comprises approximately 50 amino-acids and is similar to the type-1 repeats in TSP-1 and TSP-2 (57). The human ADAMTS family contains 19 members that can be clustered into eight different evolutionary clades, depending on their domain organisation and known functions. The aggrecanase and proteoglycanase clades contain members (ADAMTS1, 4, 5, 8, 15, 9, 20) that are involved in the processing of hyaluronan-binding chondroitin sulphate proteoglycan extracellular proteins, including versican and aggrecan. Another clade encloses pro-collagen N-propeptidases (ADAMTS2, 3, 14) that confer maturation of triple helical collagen fibrils (57).
The overall domain organisation of ADAMTS proteins can be structured into a metalloproteinase domain and an ancillary domain (Figure 1c). The metalloproteinase domain consists of a pro-domain, the catalytic metalloprotease domain and a disintegrin-like module. The catalytic domain of ADAMTS members contain a HExxHxBG(N/S)BxHD consensus motif, with B as a large non-polar residue and three histidines that coordinate the Zn2+ metal ion (Figure 1c and d) (8, 57, 58). To date, no ADAMTS protein has been identified to associate with integrins. In contrast, crystal structure data suggest that the disintegrin-like domain is an integral part of the catalytic core of ADAMTS proteins (11, 59). The composition of the ancillary domain varies between the different ADAMTS members; however, all have at least one TSR motif in common (Figure 1c). ADAMTS proteins display multiple functions during tissue development and homeostasis, and their dysregulation has been associated with various diseases. During development, members of the aggrecanase and proteoglycanase clade process the ECM component versican ensuring enough structural support on the one hand, while allowing dynamic remodelling on the other hand (57).
ADAMTS1 and ADAMTS2
The aggrecanase/proteoglycanase group member ADAMTS1 is involved in the processing of ECM components (60). Expression of ADAMTS1 positively correlated with progression of hepatic fibrosis to cirrhosis. ADAMTS1 was localised to HSCs. Interestingly, ADAMTS1 expression is elevated in activated HSCs and is associated with latency-associated peptide (LAP)-TGFβ, resulting in TGFβ activation (Figure 2c, Table 1). Consequently, a KTFR sequence-containing peptide derived from ADAMTS1 was sufficient to reduce collagen deposition in the murine carbon tetrachloride cirrhosis model (61). ADAMTS2 cleaves the pro-peptides of type I and type II pro-collagens prior to fibril formation. Its prominent role in collagen maturation has been further emphasised by the discovery of ADAMTS2 mutations in Ehlers-Danlos syndrome type VIIC (62), which is characterised by extreme skin fragility and joint laxity, among other symptoms. The impact of ADAMTS2 on liver biology has been previously shown in experimental liver disease using carbon tetrachloride (Table 1). While the initial parenchymal damage was similar, HSC activation and collagen deposition were significantly reduced in ADAMTS2-/- mice compared to appropriate controls (Figure 2c, Table 1) (63).
ADAMTS13
ADAMTS13 is the sole member of the von Willebrand Factor (vWF)-cleaving protease (vWFCP) clade. The vWF is a pro-thrombogenic glycoprotein, produced constitutively as ultra-large, multimeric protein by endothelial cells (64). After endothelial injury, vWF binds on one side to sub-endothelial collagen and on the other side to platelet glycoproteins, gpIb/IX/V, leading to platelet tethering at the injury site. Under fluid shear stress, vWF gets proteolytically processed. Thus, ADAMTS13 generates smaller, non-functional vWF fragments (Figure 2c) (65). Loss of ADAMTS13 catalytic activity, either by recessive mutations (66) or by inhibitory auto-antibodies (67), leads to the accumulation of ultra-large vWF and the formation of platelet-rich microthrombi in the micro-vasculature causing a life-threatening rare disorder called thrombotic thrombocytopenia purpura (68, 69). Low levels of ADAMTS13 are also found in patients with sepsis-induced disseminated intravascular coagulation and are associated with increased mortality (70).
ADAMTS13 expression is predominantly found in the liver but also to some extent in skeletal muscles, placenta and the lung (71). It is therefore not surprising that patients with severe alcoholic hepatitis (SAH) display reduced plasma activity of ADAMTS13 and reduced vWF proteolysis (Table 1). In SAH patients with multiorgan failure, ADAMTS13 levels were markedly reduced, resulting in increased platelet clumping and subsequent multiorgan failure (72). Similarly, patients with acute liver injury and acute liver failure displayed an imbalance between vWF and ADAMTS13 (Table 1), and low ADAMTS13 levels were associated with higher grades of encephalopathy and lower survival rates potentially linked to microthrombus formation (73).
A more detailed analysis localised ADAMTS13 expression to αSMA-positive HSCs in a patient with hepatitis C-related chronic hepatitis, suggesting a role in fibrotic liver disease (74). Accordingly, ADAMTS13 expression levels were increased not only in activated rat HSCs in vitro but also in HSCs in vivo in carbon tetrachloride-injured rats (75). Patients with liver cirrhosis displayed increasing levels of vWF with increasing severity according to Child-Pugh classification, while collagen-binding activity of vWF was decreased in these patients, partially correlating with increased ADAMTS13 activity (76). However, two other studies detected decreased ADAMTS13 levels in cirrhotic livers, linking ADAMTS13 levels to an increased platelet thrombi formation (77, 78). A very recent study demonstrated that while ADAMTS13 did not correlate with the Child-Pugh score in cirrhotic patients, low levels of ADAMTS13 were associated with portal vein thrombosis (79). In contrast, in patients with non-alcoholic fatty liver disease, the increased risk in thrombosis was not linked to aberrant ADAMTS13 levels or hyperactive haemostasis (80).
Conclusion
As summarised above, selected members of ADAM and ADAMTS proteinase families display altered expression in different liver pathologies, and a mechanistical role in the pathology has been shown for some members. However, we are still far from fully understanding the complex nature of ADAM and ADAMTS proteinases and their impact on liver physiology and pathology. Substrate analysis on a proteomics level will help to unveil how ADAM and ADAMTS proteinases regulate liver biology.
Further studies will also show if ADAM and ADAMTS expression and activity can be harnessed for diagnostic or therapeutic purposes. ADAMTS13 activity is already routinely assessed for the diagnosis of thrombotic thrombocytopenia purpura. Extended studies will determine the utility and benefit of ADAMTS13 activity measurement in the clinical management of acute and chronic liver disease. Albeit overexpression of some ADAM proteases has been shown in liver pathologies, more extended human studies, and experimental in vivo studies using targeted deletion, in particular hepatic cell types of the liver, are warranted to better understand the role of ADAM proteases in liver pathologies and targeted diagnosis and therapeutics.
There have been numerous research activities in the past to develop small molecule inhibitors for ADAM17 to treat chronic inflammatory diseases such as rheumatoid arthritis. However, the first chemical entities displayed musculoskeletal and hepatic toxicities and therefore did not proceed beyond phase I/II clinical trials (81). A recently developed dual-specific ADAM10/17 inhibitor, INCB7839 (25), is a promising candidate that is currently in a clinical trial phase I/II study (NCT02142451) for the treatment of diffuse large B-cell non-hodgkin lymphoma. In addition, very recently, the recombinant pro-domain of ADAM17 has been developed as a specific ADAM17 inhibitor in vitro and in vivo and might represent a novel strategy to specifically inhibit ADAM and ADAMTS proteinases (16). In summary, the world of ADAM and ADAMTS proteinases remains a rich but largely underexplored sea of therapeutic opportunities.
Conflict of interest
The authors report no conflict of interest with respect to research, authorship and/or publication of this article.
Acknowledgement
This work was supported by the Deutsche Forschungsgemeinschaft (DFG), Bonn [grant number SFB841; Liver inflammation: Infection, immune regulation and consequences, to D.S.-A.] and the Australian Technology Network/Deutscher Akademischer Austauschdienst [ATN-DAAD PPP to D.S.-A. and J.T.-P.]
References
- Gilgenkrantz H, Collin de l’Hortet A. Understanding liver regeneration: From mechanisms to regenerative medicine. Am J Pathol. 2018;188:1316–27. https://doi.org/10.1016/j.ajpath.2018.03.008
- Köhn-Gaone J, Gogoi-Tiwari J, Ramm GA, Olynyk JK, Tirnitz-Parker JEE. The role of liver progenitor cells during liver regeneration, fibrogenesis, and carcinogenesis. Am J Physiol. 2016;310:G143–54.
- Ruddell RG, Knight B, Tirnitz-Parker JEE, Akhurst B, Summerville L, Subramaniam VN, et al. Lymphotoxin-beta receptor signaling regulates hepatic stellate cell function and wound healing in a murine model of chronic liver injury. Hepatology. 2009;49:227–39. https://doi.org/10.1002/hep.22597
- Boulter L, Govaere O, Bird TG, Radulescu S, Ramachandran P, Pellicoro A, et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med. 2012;18:572–9. https://doi.org/10.1038/nm.2667
- Tirnitz-Parker JEE, Olynyk JK, Ramm GA. Role of TWEAK in coregulating liver progenitor cell and fibrogenic responses. Hepatology. 2014;59:1198–201. https://doi.org/10.1002/hep.26701
- Finkin S, Yuan D, Stein I, Taniguchi K, Weber A, Unger K, et al. Ectopic lymphoid structures function as microniches for tumor progenitor cells in hepatocellular carcinoma. Nat Immunol. 2015;16:1235–44. https://doi.org/10.1038/ni.3290
- Huxley-Jones J, Clarke T-K, Beck C, Toubaris G, Robertson DL, Boot-Handford RP. The evolution of the vertebrate metzincins; insights from Ciona intestinalis and Danio rerio. BMC Evol Biol. 2007;7:63. https://doi.org/10.1186/1471-2148-7-63
- Gomis-Rüth FX. Catalytic domain architecture of metzincin metalloproteases. Biol Chem. 2009;284:15353–7. https://doi.org/10.1074/jbc.R800069200
- Reiss K, Saftig P. The “a disintegrin and metalloprotease” (ADAM) family of sheddases: Physiological and cellular functions. Semin Cell Dev Biol. 2009;20:126–37. https://doi.org/10.1016/j.semcdb.2008.11.002
- Seegar TCM, Killingsworth LB, Saha N, Meyer PA, Patra D, Zimmerman B, et al. Structural basis for regulated proteolysis by the α-secretase ADAM10. Cell. 2017;171:1638–48. https://doi.org/10.1016/j.cell.2017.11.014
- Mosyak L, Georgiadis K, Shane T, Svenson K, Hebert T, McDonagh T, et al. Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5. Protein Sci. 2008;17:16–21. https://doi.org/10.1110/ps.073287008
- Düsterhöft S, Jung S, Hung C-W, Tholey A, Sönnichsen FD, Grötzinger J, et al. Membrane-proximal domain of a disintegrin and metalloprotease-17 represents the putative molecular switch of its shedding activity operated by protein-disulfide isomerase. J Am Chem Soc. 2013;135:5776–81. https://doi.org/10.1110/ps.073287008
- Jones JC, Rustagi S, Dempsey PJ. ADAM proteases and gastrointestinal function. Annu Rev Physiol. 2016;78:243–76. https://doi.org/10.1146/annurev-physiol-021014-071720
- Howard L, Maciewicz RA, Blobel CP. Cloning and characterization of ADAM28: Evidence for autocatalytic pro-domain removal and for cell surface localization of mature ADAM28. Biochem J. 2000;348 Pt 1:21–7. https://doi.org/10.1042/bj3480021
- Schlomann U, Wildeboer D, Webster A, Antropova O, Zeuschner D, Knight CG, et al. The metalloprotease disintegrin ADAM8. Processing by autocatalysis is required for proteolytic activity and cell adhesion. J Biol Chem. 2002;277:48210–19. https://doi.org/10.1074/jbc.M203355200
- Wong E, Cohen T, Romi E, Levin M, Peleg Y, Arad U, et al. Harnessing the natural inhibitory domain to control TNFα Converting Enzyme (TACE) activity in vivo. Sci Rep. 2016;6:35598. https://doi.org/10.1038/srep35598
- Maskos K, Fernandez-Catalan C, Huber R, Bourenkov GP, Bartunik H, Ellestad GA, et al. Crystal structure of the catalytic domain of human tumor necrosis factor-alpha-converting enzyme. Proc Natl Acad Sci U S A. 1998;95:3408–12. https://doi.org/10.1073/pnas.95.7.3408
- Stawikowska R, Cudic M, Giulianotti M, Houghten RA, Fields GB, Minond D. Activity of ADAM17 (a disintegrin and metalloprotease 17) is regulated by its noncatalytic domains and secondary structure of its substrates. J Biol Chem. 2013;288:22871–9. https://doi.org/10.1074/jbc.M113.462267
- Janes PW, Saha N, Barton WA, Kolev MV, Wimmer-Kleikamp SH, Nievergall E, et al. Adam meets Eph: An ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell. 2005;123:291–304. https://doi.org/10.1016/j.cell.2005.08.014
- Xu P, Derynck R. Direct activation of TACE-mediated ectodomain shedding by p38 MAP kinase regulates EGF receptor-dependent cell proliferation. Mol Cell. 2010;37:551–66. https://doi.org/10.1016/j.molcel.2010.01.034
- Xu P, Liu J, Sakaki-Yumoto M, Derynck R. TACE activation by MAPK-mediated regulation of cell surface dimerization and TIMP3 association. Sci Signal. 2012;5:ra34. https://doi.org/10.1126/scisignal.2002689
- Liu C, Xu P, Lamouille S, Xu J, Derynck R. TACE-mediated ectodomain shedding of the type I TGF-beta receptor downregulates TGF-beta signaling. Mol Cell. 2009;35:26–36. https://doi.org/10.1016/j.molcel.2009.06.018
- Scheller J, Chalaris A, Garbers C, Rose-John S. ADAM17: A molecular switch to control inflammation and tissue regeneration Trends Immunol. 2011;32:380–7. https://doi.org/10.1016/j.it.2011.05.005
- Schmidt-Arras D, Rose-John S. IL-6 pathway in the liver: From physiopathology to therapy. J Hepatol. 2016;64:1403–15. https://doi.org/10.1016/j.jhep.2016.02.004
- Moss ML, Minond D. Recent advances in ADAM17 research: A promising target for cancer and inflammation. Mediators Inflamm. 2017;2017:9673537.
- Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, et al. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet. 2002;11:2615–24. https://doi.org/10.1093/hmg/11.21.2615
- White JM. ADAMs: Modulators of cell-cell and cell-matrix interactions. Curr Opin Cell Biol. 2003;15:598–606. https://doi.org/10.1016/j.ceb.2003.08.001
- Oria VO, Lopatta P, Schilling O. The pleiotropic roles of ADAM9 in the biology of solid tumors. Cell Mol Life Sci. 2018;75:2291–301. https://doi.org/10.1007/s00018-018-2796-x
- Schwettmann L, Wehmeier M, Jokovic D, Aleksandrova K, Brand K, Manns MP, et al. Hepatic expression of A disintegrin and metalloproteinase (ADAM) and ADAMs with thrombospondin motives (ADAM-TS) enzymes in patients with chronic liver diseases. J Hepatol. 2008;49:243–50. https://doi.org/10.1016/j.jhep.2008.03.020
- Le Pabic H, Bonnier D, Wewer UM, Coutand A, Musso O, Baffet G, et al. ADAM12 in human liver cancers: TGF-beta-regulated expression in stellate cells is associated with matrix remodeling. Hepatology. 2003;37:1056–66. https://doi.org/10.1053/jhep.2003.50205
- Tannapfel A, Anhalt K, Häusermann P, Sommerer F, Benicke M, Uhlmann D, et al. Identification of novel proteins associated with hepatocellular carcinomas using protein microarrays. J Pathol. 2003;201:238–49. https://doi.org/10.1002/path.1420
- Kohga K, Takehara T, Tatsumi T, Ishida H, Miyagi T, Hosui A, et al. Sorafenib inhibits the shedding of major histocompatibility complex class I-related chain A on hepatocellular carcinoma cells by down-regulating a disintegrin and metalloproteinase 9. Hepatology. 2010;51:1264–73. https://doi.org/10.1002/hep.23456
- Mazzocca A, Coppari R, De Franco R, Cho J-Y, Libermann TA, Pinzani M, et al. A secreted form of ADAM9 promotes carcinoma invasion through tumor-stromal interactions. Cancer Res. 2005;65:4728–38. https://doi.org/10.1158/0008-5472.CAN-04-4449
- Geisler F, Nagl F, Mazur PK, Lee M, Zimber-Strobl U, Strobl LJ, et al. Liver-specific inactivation of Notch2, but not Notch1, compromises intrahepatic bile duct development in mice. Hepatology. 2008;48:607–16. https://doi.org/10.1002/hep.22381
- Sparks EE, Huppert KA, Brown MA, Washington MK, Huppert SS. Notch signaling regulates formation of the three-dimensional architecture of intrahepatic bile ducts in mice. Hepatology. 2010;51:1391–400. https://doi.org/10.1002/hep.23431
- Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A et al., Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235–42. https://doi.org/10.1038/ng0797-235
- Müller M, Wetzel S, Köhn-Gaone J, Chalupsky K, Lüllmann-Rauch R, Barikbin R, et al. A disintegrin and metalloprotease 10 (ADAM10) is a central regulator of murine liver tissue homeostasis. Oncotarget. 2016;7:17431–41. https://doi.org/10.18632/oncotarget.7836
- Bourd-Boittin K, Basset L, Bonnier D, L’helgoualc’h A, Samson M, Théret N. CX3CL1/fractalkine shedding by human hepatic stellate cells: Contribution to chronic inflammation in the liver. J Cell Mol Med. 2009;13:1526–35. https://doi.org/10.1111/j.1582-4934.2009.00787.x
- Kohga K, Takehara T, Tatsumi T, Miyagi T, Ishida H, Ohkawa K, et al. Anticancer chemotherapy inhibits MHC class I-related chain a ectodomain shedding by downregulating ADAM10 expression in hepatocellular carcinoma. Cancer Res. 2009;69:8050–7. https://doi.org/10.1158/0008-5472.CAN-09-0789
- Yuan S, Lei S, Wu S. ADAM10 is overexpressed in human hepatocellular carcinoma and contributes to the proliferation, invasion and migration of HepG2 cells. Oncol Rep. 2013;30:1715–22. https://doi.org/10.3892/or.2013.2650
- Chalupský K, Kanchev I, Žbodáková O, Buryová H, Jiroušková M, Kořínek V, et al. ADAM10/17-dependent release of soluble c-Met correlates with hepatocellular damage. Folia Biol (Praha) 2013;59:76–86.
- Zhang Y-W, Graveel C, Shinomiya N, Vande Woude GF. Met decoys: Will cancer take the bait? Cancer Cell. 2004;6:5–6. https://doi.org/10.1016/j.ccr.2004.07.003
- Gavert N, Sheffer M, Raveh S, Spaderna S, Shtutman M, Brabletz T, et al. Expression of L1-CAM and ADAM10 in human colon cancer cells induces metastasis. Cancer Res. 2007;67:7703–12. https://doi.org/10.1158/0008-5472.CAN-07-0991
- Kopitz C, Gerg M, Bandapalli OR, Ister D, Pennington CJ, Hauser S, et al. Tissue inhibitor of metalloproteinases-1 promotes liver metastasis by induction of hepatocyte growth factor signaling. Cancer Res. 2007;67:8615–23. https://doi.org/10.1158/0008-5472.CAN-07-0232
- Schelter F, Grandl M, Seubert B, Schaten S, Hauser S, Gerg M, et al. Tumor cell-derived Timp-1 is necessary for maintaining metastasis-promoting Met-signaling via inhibition of Adam-10. Clin Exp Metastasis. 2011;28:793–802. https://doi.org/10.1007/s10585-011-9410-z
- Grünwald B, Harant V, Schaten S, Frühschütz M, Spallek R, Höchst B et al., Pancreatic premalignant lesions secrete tissue inhibitor of metalloproteinases-1, which activates hepatic stellate cells via CD63 signaling to create a premetastatic niche in the liver. Gastroenterology. 2016;151:1011–24.e7. https://doi.org/10.1053/j.gastro.2016.07.043
- Nyren-Erickson EK, Jones JM, Srivastava D, Mallik S. A disintegrin and metalloproteinase-12 ADAM12): Function, roles in disease progression, and clinical implications Biochim Biophys Acta. 2013;1830:4445–55.
- Daduang J, Limpaiboon T, Daduang S. Biomarker to distinguish hepatocellular carcinoma from cholangiocarcinoma by serum a disintegrin and metalloprotease 12. Arch Med Sci. 2011;7:1013–16. https://doi.org/10.5114/aoms.2011.26613
- Fang T, Lin J, Wang Y, Chen G, Huang J, Chen J, et al. Tetraspanin-8 promotes hepatocellular carcinoma metastasis by increasing ADAM12m expression. Oncotarget. 2016;7:40630–43. https://doi.org/10.18632/oncotarget.9769
- He G, Dhar D, Nakagawa H, Font-Burgada J, Ogata H, Jiang Y, et al. Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell. 2013;155:384–96. https://doi.org/10.1016/j.cell.2013.09.031
- Argast GM, Campbell JS, Brooling JT, Fausto N. Epidermal growth factor receptor transactivation mediates tumor necrosis factor-induced hepatocyte replication. J Biol Chem. 2004;279:34530–6. https://doi.org/10.1074/jbc.M405703200
- Murillo MM, del Castillo G, Sánchez A, Fernández M, Fabregat I. Involvement of EGF receptor and c-Src in the survival signals induced by TGF-beta1 in hepatocytes. Oncogene. 2005;24:4580–7. https://doi.org/10.1038/sj.onc.1208664
- Li Y, Ren Z, Wang Y, Dang Y-Z, Meng B-X, Wang G-D, et al. ADAM17 promotes cell migration and invasion through the integrin β1 pathway in hepatocellular carcinoma. Exp Cell Res. 2018;370:373–82. https://doi.org/10.1016/j.yexcr.2018.06.039
- Wang R, Li Y, Tsung A, Huang H, Du Q, Yang M, et al. iNOS promotes CD24+, CD133+ liver cancer stem cell phenotype through a TACE/ADAM17-dependent Notch signaling pathway. Proc Natl Acad Sci U S A. 2018;115:E10127–36. https://doi.org/10.1073/pnas.1722100115
- Hong SW, Hur W, Choi JE, Kim J-H, Hwang D, Yoon SK. Role of ADAM17 in invasion and migration of CD133-expressing liver cancer stem cells after irradiation. Oncotarget. 2016;7:23482–97. https://doi.org/10.18632/oncotarget.8112
- Lu J, Ye X, Fan F, Xia L, Bhattacharya R, Bellister S, et al. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell. 2013;23:171–85.
https://doi.org/10.1016/j.ccr.2012.12.021
- Bergmann J, Müller M, Baumann N, Reichert M, Heneweer C, Bolik J, et al. IL-6 trans-signaling is essential for the development of hepatocellular carcinoma in mice. Hepatology. 2017;65:89–103. https://doi.org/10.1002/hep.28874
- Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) family. Genome Biol. 2015;16:113. https://doi.org/10.1186/s13059-015-0676-3
- Stanton H, Melrose J, Little CB, Fosang AJ. Proteoglycan degradation by the ADAMTS family of proteinases. Biochim Biophys Acta. 2011;1812:1616–29. https://doi.org/10.1016/j.bbadis.2011.08.009
- Gerhardt S, Hassall G, Hawtin P, McCall E, Flavell L, Minshull C, et al. Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains. J Mol Biol. 2007;373:891–902. https://doi.org/10.1016/j.jmb.2007.07.047
- Bourd-Boittin K, Bonnier D, Leyme A, Mari B, Tuffery P, Samson M, et al. Protease profiling of liver fibrosis reveals the ADAM metallopeptidase with thrombospondin type 1 motif, 1 as a central activator of transforming growth factor beta. Hepatology. 2011;54:2173–84. https://doi.org/10.1002/hep.24598
- Colige A, Sieron AL, Li SW, Schwarze U, Petty E, Wertelecki W, et al. Human Ehlers-Danlos syndrome type VII C and bovine dermatosparaxis are caused by mutations in the procollagen I N-proteinase gene. Am J Hum Genet. 1999;65:308–17. https://doi.org/10.1086/302504
- Kesteloot F, Desmoulière A, Leclercq I, Thiry M, Arrese JE, Prockop DJ, et al. ADAM metallopeptidase with thrombospondin type 1 motif 2 inactivation reduces the extent and stability of carbon tetrachloride-induced hepatic fibrosis in mice. Hepatology. 2007;46:1620–31. https://doi.org/10.1002/hep.21868
- Sadler JE. von Willebrand factor assembly and secretion. J Thromb Haemost. 2009;7 Suppl 1:24–7. https://doi.org/10.1111/j.1538-7836.2009.03375.x
- Sadler JE. Von Willebrand factor, ADAMTS13, and thrombotic thrombocytopenic purpura. Blood. 2008;112:11–18. https://doi.org/10.1182/blood-2008-02-078170
- Levy GG, Nichols WC, Lian EC, Foroud T, McClintick JN, McGee BM, et al. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature. 2001;413:488–94. https://doi.org/10.1038/35097008
- Zheng XL. ADAMTS13 and von Willebrand factor in thrombotic thrombocytopenic purpura. Annu Rev Med. 2015;66:211–25.
- Bianchi V, Robles R, Alberio L, Furlan M, Lämmle B. Von Willebrand factor-cleaving protease ADAMTS13) in thrombocytopenic disorders: A severely deficient activity is specific for thrombotic thrombocytopenic purpura. Blood. 2002;100:710–13. https://doi.org/10.1182/blood-2002-02-0344
- Ono T, Mimuro J, Madoiwa S, Soejima K, Kashiwakura Y, Ishiwata A, et al. Severe secondary deficiency of von Willebrand factor-cleaving protease (ADAMTS13) in patients with sepsis-induced disseminated intravascular coagulation: Its correlation with development of renal failure. Blood. 2006;107:528–34. https://doi.org/10.1182/blood-2005-03-1087
- Azfar MF, Khan MF, Habib SS, Aseri ZA, Zubaidi AM, Aguila DO, et al. Prognostic value of ADAMTS13 in patients with severe sepsis and septic shock. Clin Invest Med. 2017;40:E49–58. https://doi.org/10.25011/cim.v40i2.28195
- Levy GG, Motto DG, Ginsburg D. ADAMTS13 turns 3. Blood. 2005;106:11–17. https://doi.org/10.1182/blood-2004-10-4097
- Uemura M, Matsuyama T, Ishikawa M, Fujimoto M, Kojima H, Sakurai S, et al. Decreased activity of plasma ADAMTS13 may contribute to the development of liver disturbance and multiorgan failure in patients with alcoholic hepatitis. Alcohol Clin Exp Res. 2005;29:264S–271S.
- Hugenholtz GCG, Adelmeijer J, Meijers JCM, Porte RJ, Stravitz RT, Lisman T. An unbalance between von Willebrand factor and ADAMTS13 in acute liver failure: Implications for hemostasis and clinical outcome. Hepatology. 2013;58:752–61. https://doi.org/10.1002/hep.26372
- Uemura M, Tatsumi K, Matsumoto M, Fujimoto M, Matsuyama T, Ishikawa M, et al. Localization of ADAMTS13 to the stellate cells of human liver. Blood. 2005;106:922–4. https://doi.org/10.1182/blood-2005-01-0152
- Niiya M, Uemura M, Zheng XW, Pollak ES, Dockal M, Scheiflinger F, et al. Increased ADAMTS-13 proteolytic activity in rat hepatic stellate cells upon activation in vitro and in vivo. J Thromb Haemost. 2006;4:1063–70. https://doi.org/10.1111/j.1538-7836.2006.01893.x
- Lisman T, Bongers TN, Adelmeijer J, Janssen HLA, de Maat MPM, de Groot PG, et al. Elevated levels of von Willebrand factor in cirrhosis support platelet adhesion despite reduced functional capacity. Hepatology. 2006;44:53–61. https://doi.org/10.1002/hep.21231
- Mannucci PM, Canciani MT, Forza I, Lussana F, Lattuada A, Rossi E. Changes in health and disease of the metalloprotease that cleaves von Willebrand factor. Blood. 2001;98:2730–5. https://doi.org/10.1182/blood.V98.9.2730
- Uemura M, Fujimura Y, Matsumoto M, Ishizashi H, Kato S, Matsuyama T, et al. Comprehensive analysis of ADAMTS13 in patients with liver cirrhosis. Thromb Haemost. 2008;99:1019–29. https://doi.org/10.1160/TH08-01-0006
- Lancellotti S, Basso M, Veca V, Sacco M, Riccardi L, Pompili M, et al. Presence of portal vein thrombosis in liver cirrhosis is strongly associated with low levels of ADAMTS-13: A pilot study. Intern Emerg Med. 2016;11:959–67. https://doi.org/10.1007/s11739-016-1467-x
- Potze W, Siddiqui MS, Boyett SL, Adelmeijer J, Daita K, Sanyal AJ, et al. Preserved hemostatic status in patients with non-alcoholic fatty liver disease. J Hepatol. 2016;65:980–7. https://doi.org/10.1016/j.jhep.2016.06.001
- Moss ML, Sklair-Tavron L, Nudelman R. Drug insight: Tumor necrosis factor-converting enzyme as a pharmaceutical target for rheumatoid arthritis Nat Clin Pract Rheumatol. 2008;4:300–9. https://doi.org/10.1038/ncprheum0797